LASE is a software dedicated to the X-Ray Absorption spectra analysis. It can run on any computer with unix and X-Window, or with Windows (95 or higher). In both case, it offers a userfriendly graphical interface, which allows to visualize in real time the effects of various parameters, while offering a great liberty to the user.
LASE was first designed at LURE, for the analysis of bioinorganical samples studied by X-ray absorption spectroscopy. Hence, it is especially suited for this kind of samples: proteins, transition metals complexes,...
Currently, LASE offers tools for analysis and treatment of spectra in three main categories:
These points are presented with more details hereafter.
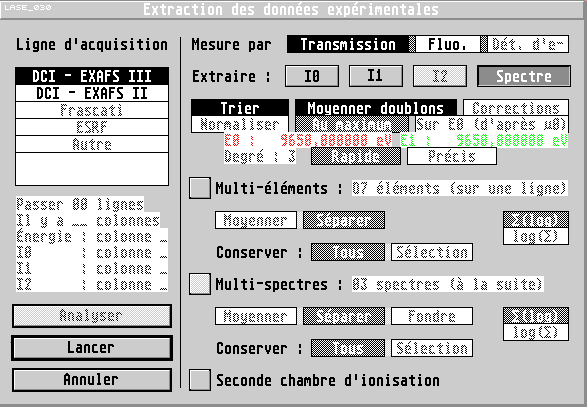
LASE offers various tools to treat the experimental data and extracting the EXAFS oscillations. Especially, it also includes statistical tools to handle the informations coming from the repeated measurements of a sample spectrum.
It is done with the classical Fourier transformation procedure.
LASE offers tools to help in the creation of the structural model needed to compute the theoretical EXAFS spectra that will be used in the fitting step. Presently, these models can be used for creating FEFF input files.
For its models, LASE works with "molecules": a molecule is a set of atoms grouped for a logical reason: molecule in the chemical meaning, but also unit cell(s) or other association. In that molecule one or more atoms can be defined as "absorbing": an absorbing atom represent the atom studied with the X-ray spectroscopy and regroups a set of models (including scattering paths for electrons) and experimental data.
It is possible to load in LASE structural data from the *.cor files (Cambridge Cristallographic Databank), the *.pdb files (Protein Data Bank) and simple free-format ASCII files. With the help of the babel software, that can convert various formats into the pdb file format, it is possible to load practically any structural file in LASE.
A structural model can also be created directly in LASE, starting with orthonormal coordinates or fractionnar cristallographic coordinates. For some space groups (P1, P-1, P21, C2/c, Pnma,...), symmetries are handled. Several cells can be generated at once.
If OpenGL is present (MESA
works quite well), one can view in LASE the structural model
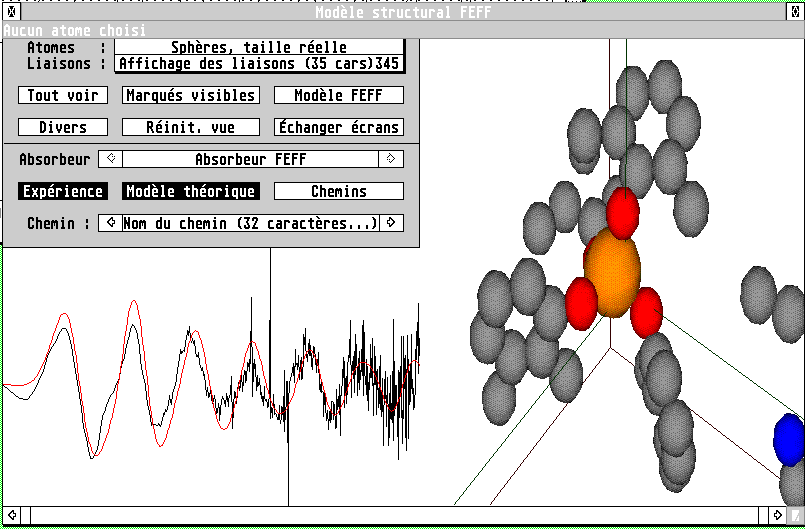
It is then possible to know atomic coordinates, distances and angles between atoms simply by clicking them. It is also possible to add or remove atoms to the FEFF model in the same way. If there is an absorber atom with a model, the experimental and modelled spectra are also shown. The various scattering paths, and their contribution to the EXAFS signal after fitting, can also be drawn.
LASE offers a graphical user interface to parameter almost all FEFF-6 options, except the OVERLAP one.
The structural model creation can be done automatically starting with the structural informations created in LASE, including all atoms or atoms closer than a given distance of the absorbing atom. The model can then be edited using the above dialog boxes.
With LASE, it is possible to fit in k-space (that is, EXAFS oscillations) with or without error-bars weighting. This fit relies on the path-decomposition of the EXAFS formula (in the case of just single scattering is present, a path is equivalent to a reflector atoms shell).
For each path, it is possible to fit its degeneracy (N, number of neighbours), Debye-Waller factor, length (R, distance) and four cumulants accounting for asymetries in statistical or thermal disorder. If required, one can also fit individual mean-free-path and edge-energy corrections.
Are also fitted a global edge-energy correction and the electronic amplitude reduction factor (S02).
While fitting, the fitted spectrum is redrawn in real time, to follow on screen the minimisation procedure.
LASE can read paths.dat files created by FEFF to construct the structural model (if not already known), the various reflexion paths and their phases and amplitudes.
LASE offers tools for performing a Monte-Carlo study of the fitting results, to obtain the average value and error bars on the fitted parameters, with no other hypothesis than the gaussian distribution of the experimental errors.